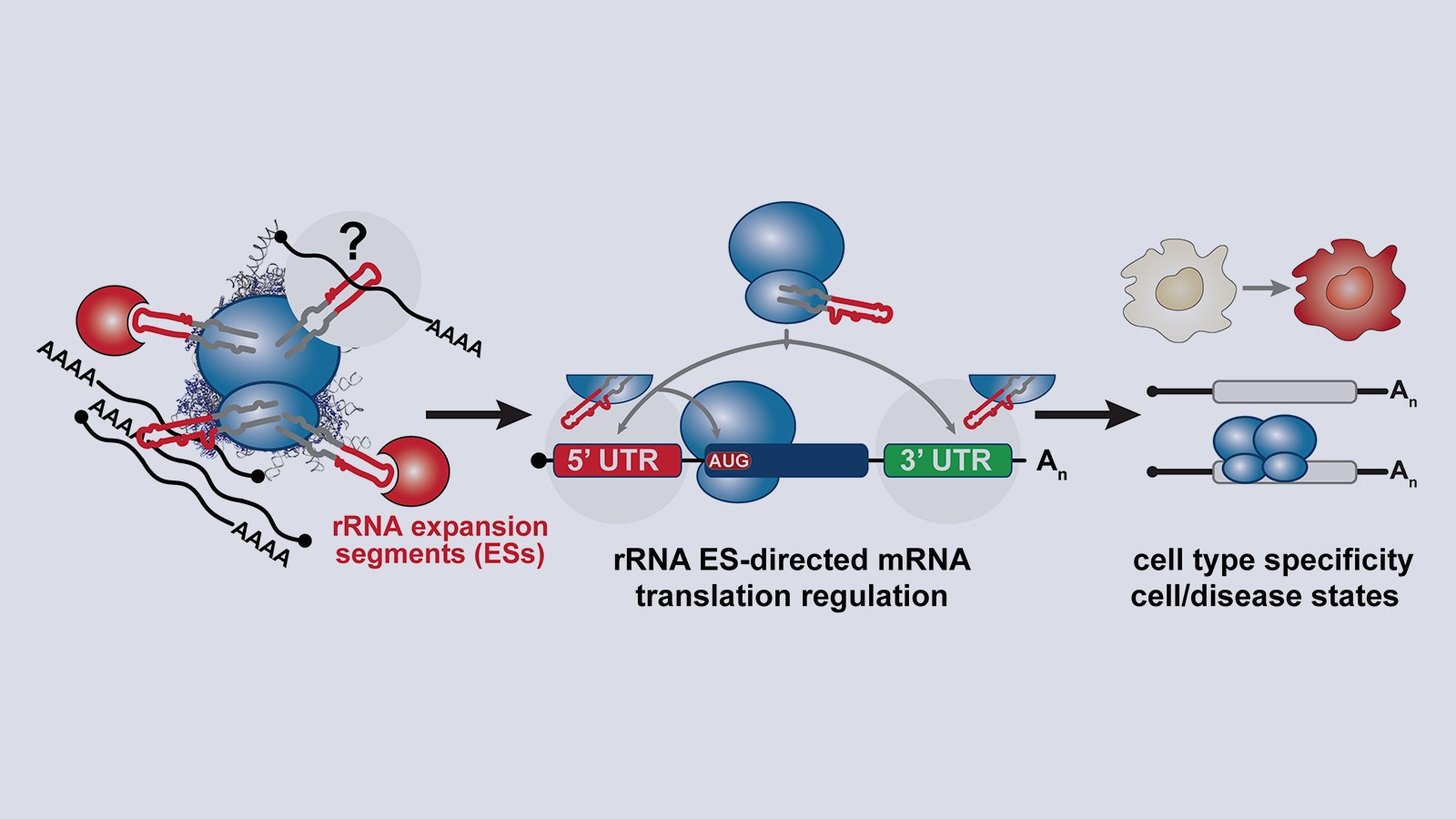
Ribosome size has dramatically increased across eukaryotic evolution, due in part to sequence insertions called rRNA expansion segments (ESs) that “expand” rRNA on the outer ribosome shell. rRNA ESs neither contribute to nor interfere with rRNA’s essential role in peptide-bond formation, so why do they exist? And what do they do? Since the 1970s, translation initiation by direct binding of rRNA and mRNA to start translation has been a classic paradigm in prokaryotes, but such evidence in eukaryotes is sparse. Kathrin’s postdoctoral work revealed a mode of gene regulation by which rRNAs, particularly an exposed rRNA ES, directly bind specific transcripts to control their translation.
Thereby, rRNA itself is a critical regulator of gene expression where rRNA ESs on the outer ribosome directly recruit specific subsets of mRNAs for selective translation. The eukaryotic ribosome contains a multitude of distinct ESs of different sequence and structure. What other functions rRNA ESs may have in gene expression regulation remains enigmatic.
A central question is: How does the ribosome recognize specific mRNAs for selective translation?