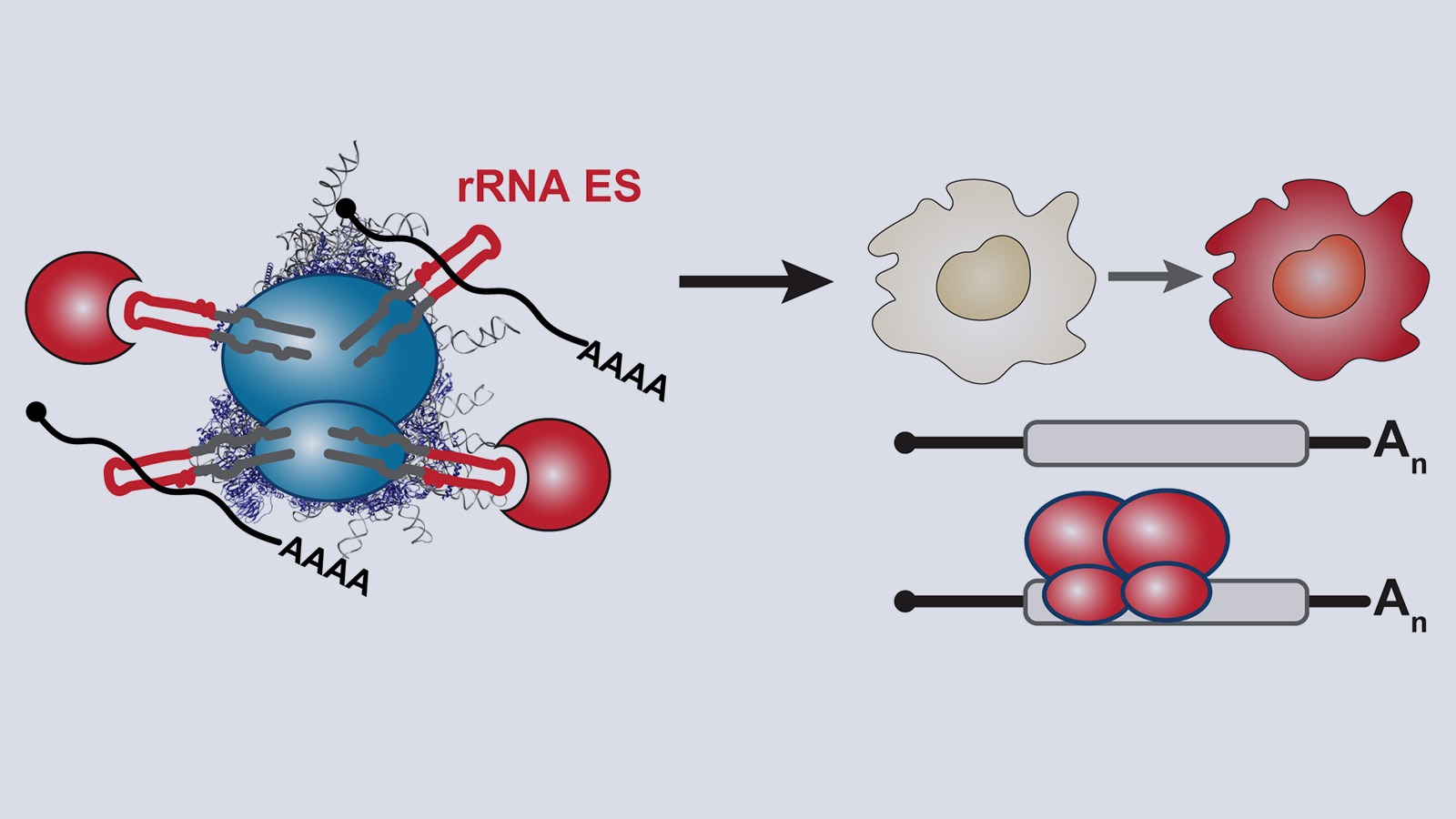
Transcript-specific regulation by the ribosome may be important in biological systems that require rapid gene expression responses to change cellular morphology and behavior. Cells that rapidly and reversibly remodel their expression patterns often rely on versatile posttranscriptional mechanisms. Kathrin’s PhD work revealed a new layer of regulation governing how macrophages rapidly change their expression patterns upon bacterial infection, which is vital for host survival. That work unraveled how a conserved class of short RNA stem-loops in the 3’ UTR is bound by the RNA-binding protein Roquin to induce rapid decay of the mRNA of many immune regulators, including tumor necrosis factor (TNF) – the most potent pro-inflammatory human cytokine secreted by macrophages and the cause of septic shock (Leppek et al., Cell, 2013).
We explore the role of ribosome-regulation of selective translation in shaping the rapid innate immune response upon bacterial infections. We focus on understanding the functional components of ribosome-mRNA contacts in a model of macrophage stimulation, where customized translation and rapid remodeling of the proteome may underlie dynamic cellular specification. Thus, we view the innate immune response of macrophages though the lens of ribosome-mediated gene regulation and aim to uncover the molecular components that orchestrate these complex and dynamic regulatory mechanisms at the level of translation.
A central question is: What is the biological impact of ribosome-directed translation in regulating key survival processes such as innate immunity?