How does rRNA-directed transcript- and species-specific mRNA translation by the ribosome regulate protein synthesis in the innate immune response?
Our lab aims to understand how the ribosome regulates species-specific, selective translation and how these principles are employed in innate immunity. We combine the disciplines of RNA biochemistry and ribosome biology with innate immunity to understand detailed molecular mechanisms in health and disease. We also develop RNA-centric tools that enable the investigation of RNA design principles for therapeutic purposes.
To read more about our research, see our research and publications.
Regulation of gene expression decides which proteins to make under which conditions and is therefore essential to forming specific cell types and tissues. Our lab studies how the interactions between ribosomes and mRNAs executes such regulation, particularly in context of the innate immune response.
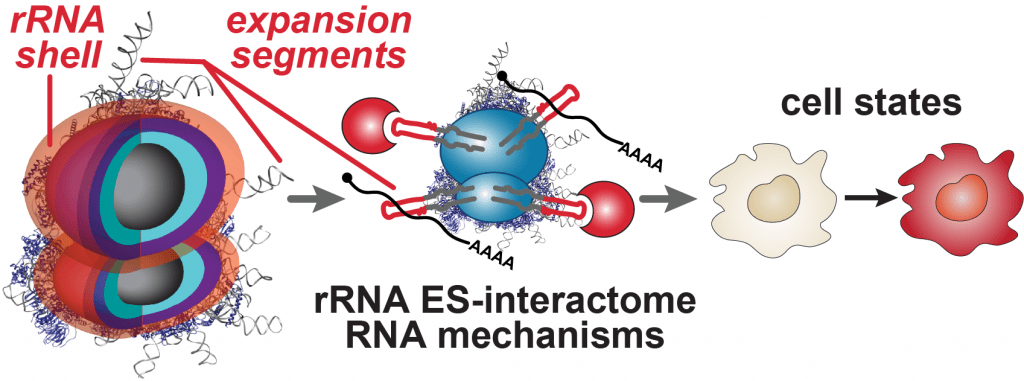
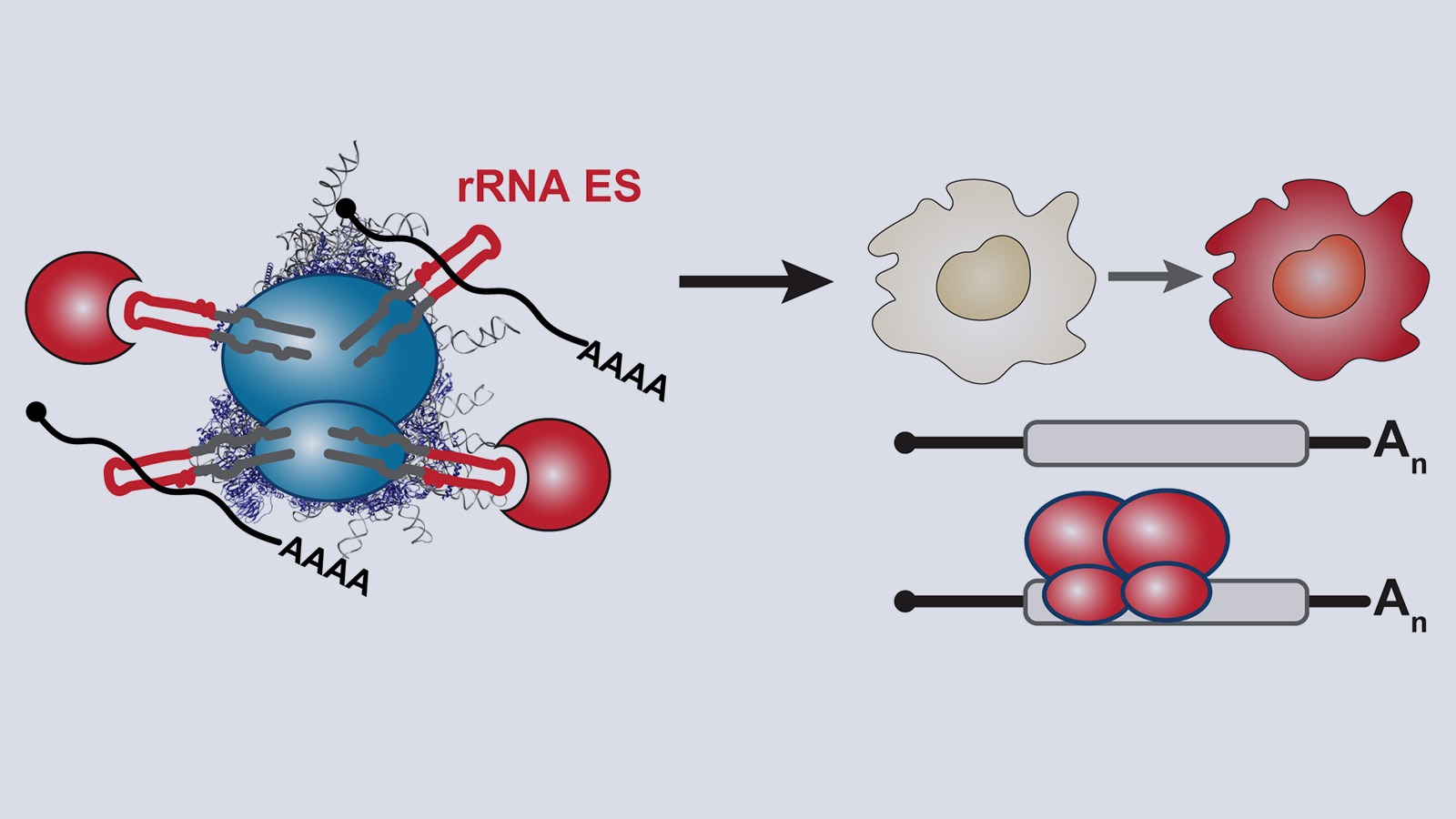
The ribosome, one of life’s most ancient molecular machines, was recently found to be an active regulator of gene expression. Ribosomes are not all identical in composition and do not translate all mRNAs equally; so-called “specialized” ribosomes preferentially translate certain transcripts to diversify gene expression. We call this process “selective translation”. It is poorly understood how ribosomal RNAs (rRNAs) mediate such selectivity. The regulatory capacity of rRNAs, some of the oldest molecules on earth, has long remained unexplored.
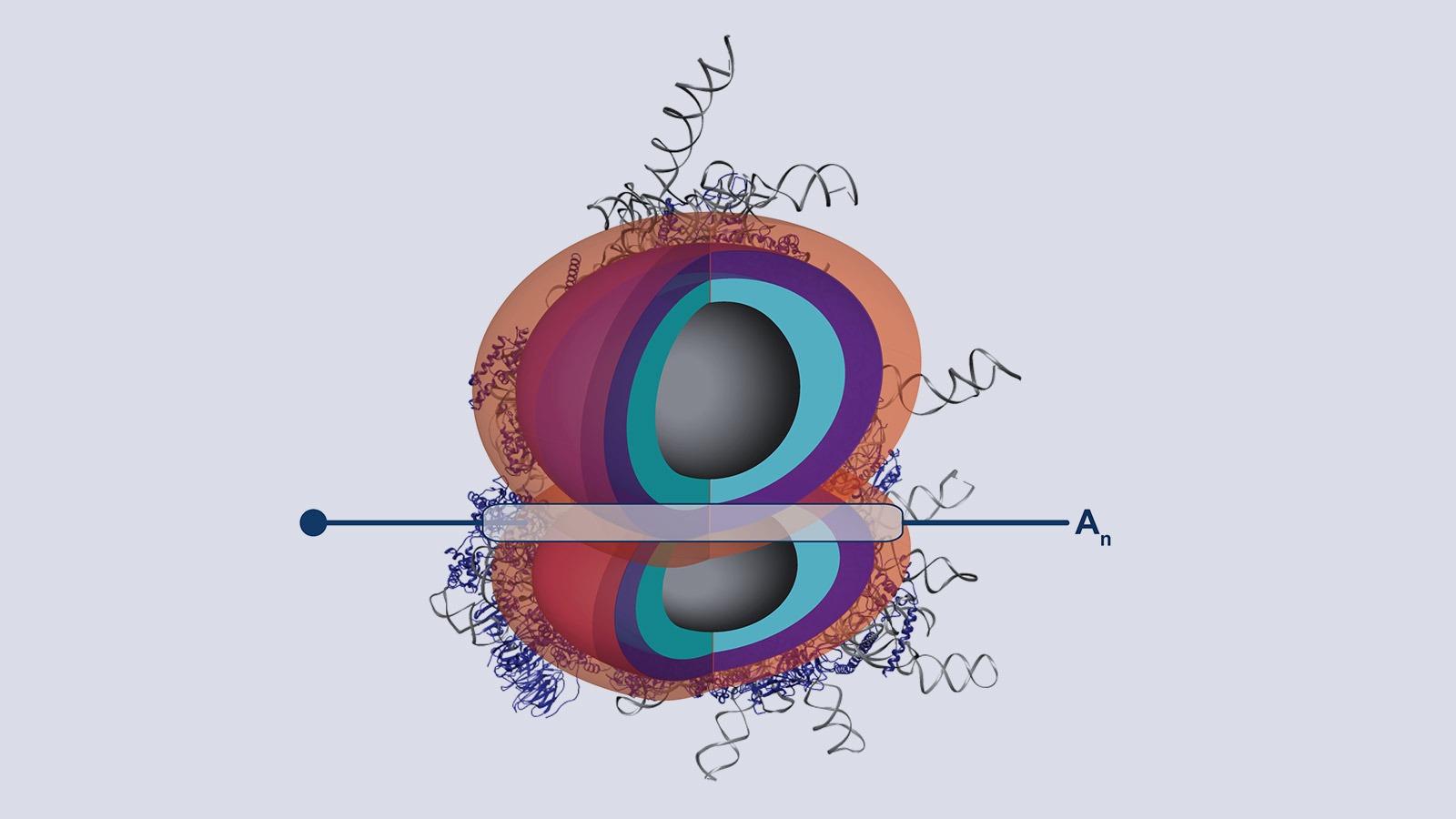
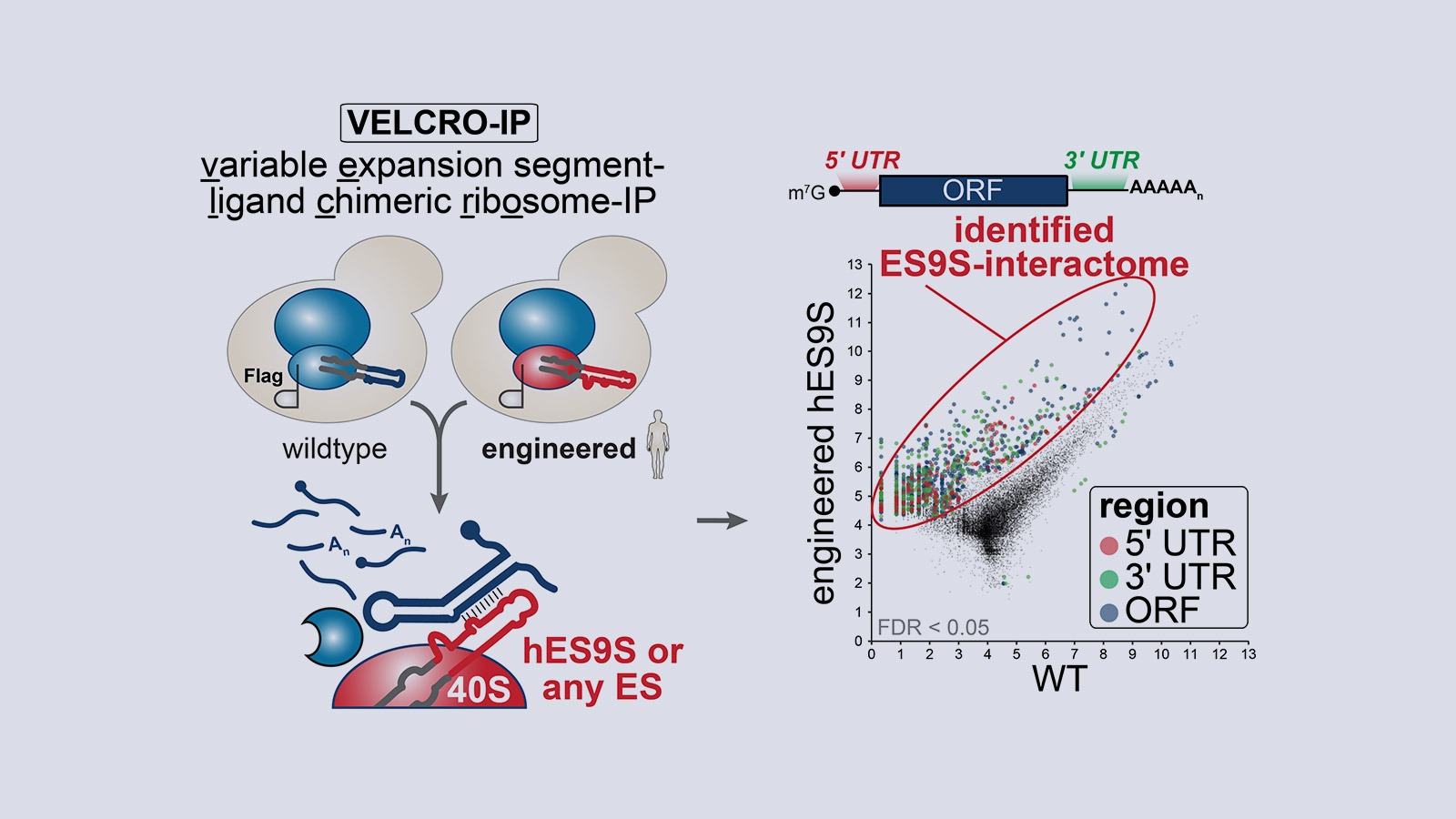
Our lab studies a fundamentally new mode of gene regulation by which rRNA regions exposed on the outer shell of the ribosome, called rRNA expansion segments (ES), directly bind to specific transcripts to control selective species-specific translation that diversifies the proteome (Leppek et al., 2020; Leppek, Byeon et al., 2021).